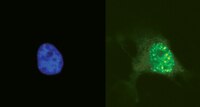
TODRA, a lncRNA at the RAD51 Locus, Is Oppositely Regulated to RAD51, and Enhances RAD51-Dependent DSB (Double Strand Break) Repair.
Gazy, I; Zeevi, DA; Renbaum, P; Zeligson, S; Eini, L; Bashari, D; Smith, Y; Lahad, A; Goldberg, M; Ginsberg, D; Levy-Lahad, E
PLoS One
10
e0134120
2015
Abstract anzeigen
Expression of RAD51, a crucial player in homologous recombination (HR) and DNA double-strand break (DSB) repair, is dysregulated in human tumors, and can contribute to genomic instability and tumor progression. To further understand RAD51 regulation we functionally characterized a long non-coding (lnc) RNA, dubbed TODRA (Transcribed in the Opposite Direction of RAD51), transcribed 69bp upstream to RAD51, in the opposite direction. We demonstrate that TODRA is an expressed transcript and that the RAD51 promoter region is bidirectional, supporting TODRA expression (7-fold higher than RAD51 in this assay, p = 0.003). TODRA overexpression in HeLa cells induced expression of TPIP, a member of the TPTE family which includes PTEN. Similar to PTEN, we found that TPIP co-activates E2F1 induction of RAD51. Analysis of E2F1's effect on the bidirectional promoter showed that E2F1 binding to the same site that promotes RAD51 expression, results in downregulation of TODRA. Moreover, TODRA overexpression induces HR in a RAD51-dependent DSB repair assay, and increases formation of DNA damage-induced RAD51-positive foci. Importantly, gene expression in breast tumors supports our finding that E2F1 oppositely regulates RAD51 and TODRA: increased RAD51 expression, which is associated with an aggressive tumor phenotype (e.g. negative correlation with positive ER (r = -0.22, p = 0.02) and positive PR status (r = -0.27, p<0.001); positive correlation with ki67 status (r = 0.36, p = 0.005) and HER2 amplification (r = 0.41, p = 0.001)), correlates as expected with lower TODRA and higher E2F1 expression. However, although E2F1 induction resulted in TPIP downregulation in cell lines, we find that TPIP expression in tumors is not reduced despite higher E2F1 expression, perhaps contributing to increased RAD51 expression. Our results identify TPIP as a novel E2F1 co-activator, suggest a similar role for other TPTEs, and indicate that the TODRA lncRNA affects RAD51 dysregulation and RAD51-dependent DSB repair in malignancy. Importantly, gene expression in breast tumors supports our finding that E2F1 oppositely regulates RAD51 and TODRA: increased RAD51 expression, which is associated with an aggressive tumor phenotype (e.g. negative correlation with positive ER (r = -0.22, p = 0.02) and positive PR status (r = -0.27, p<0.001); positive correlation with ki67 status (r = 0.36, p = 0.005) and HER2 amplification (r = 0.41, p = 0.001)), correlates as expected with lower TODRA and higher E2F1 expression. However, although E2F1 induction resulted in TPIP downregulation in cell lines, we find that TPIP expression in tumors is not reduced despite higher E2F1 expression, perhaps contributing to increased RAD51 expression. Our results identify TPIP as a novel E2F1 co-activator, suggest a similar role for other TPTEs, and indicate that the TODRA lncRNA affects RAD51 dysregulation and RAD51-dependent DSB repair in malignancy. | | | 26230935
 |
Truncating mutation in the autophagy gene UVRAG confers oncogenic properties and chemosensitivity in colorectal cancers.
He, S; Zhao, Z; Yang, Y; O'Connell, D; Zhang, X; Oh, S; Ma, B; Lee, JH; Zhang, T; Varghese, B; Yip, J; Dolatshahi Pirooz, S; Li, M; Zhang, Y; Li, GM; Ellen Martin, S; Machida, K; Liang, C
Nature communications
6
7839
2015
Abstract anzeigen
Autophagy-related factors are implicated in metabolic adaptation and cancer metastasis. However, the role of autophagy factors in cancer progression and their effect in treatment response remain largely elusive. Recent studies have shown that UVRAG, a key autophagic tumour suppressor, is mutated in common human cancers. Here we demonstrate that the cancer-related UVRAG frameshift (FS), which does not result in a null mutation, is expressed as a truncated UVRAG(FS) in colorectal cancer (CRC) with microsatellite instability (MSI), and promotes tumorigenesis. UVRAG(FS) abrogates the normal functions of UVRAG, including autophagy, in a dominant-negative manner. Furthermore, expression of UVRAG(FS) can trigger CRC metastatic spread through Rac1 activation and epithelial-to-mesenchymal transition, independently of autophagy. Interestingly, UVRAG(FS) expression renders cells more sensitive to standard chemotherapy regimen due to a DNA repair defect. These results identify UVRAG as a new MSI target gene and provide a mechanism for UVRAG participation in CRC pathogenesis and treatment response. | | | 26234763
 |
In vitro engineering of human 3D chondrosarcoma: a preclinical model relevant for investigations of radiation quality impact.
Hamdi, DH; Barbieri, S; Chevalier, F; Groetz, JE; Legendre, F; Demoor, M; Galera, P; Lefaix, JL; Saintigny, Y
BMC cancer
15
579
2015
Abstract anzeigen
The benefit of better ballistic and higher efficiency of carbon ions for cancer treatment (hadron-therapy) is asserted since decades, especially for unresectable or resistant tumors like sarcomas. However, hadron-therapy with carbon ions stays underused and raises some concerns about potential side effects for patients. Chondrosarcoma is a cartilaginous tumor, chemo- and radiation-resistant, that lacks reference models for basic and pre-clinical studies in radiation-biology. Most studies about cellular effects of ionizing radiation, including hadrons, were performed under growth conditions dramatically different from human homeostasis. Tridimensional in vitro models are a fair alternative to animal models to approach tissue and tumors microenvironment.By using a collagen matrix, standardized culture conditions, physiological oxygen tension and a well defined chondrosarcoma cell line, we developed a pertinent in vitro 3D model for hadron-biology studies. Low- and high-Linear Energy Transfer (LET) ionizing radiations from GANIL facilities of ~1 keV/μm and 103 ± 4 keV/μm were used respectively, at 2 Gy single dose. The impact of radiation quality on chondrosarcoma cells cultivated in 3D was analyzed on cell death, cell proliferation and DNA repair.A fair distribution of chondrosarcoma cells was observed in the whole 3D scaffold. Moreover, LET distribution in depth, for ions, was calculated and found acceptable for radiation-biology studies using this kind of scaffold. No difference in cell toxicity was observed between low- and high-LET radiations but a higher rate of proliferation was displayed following high-LET irradiation. Furthermore, 3D models presented a higher and longer induction of H2AX phosphorylation after 2 Gy of high-LET compared to low-LET radiations.The presented results show the feasibility and usefulness of our 3D chondrosarcoma model in the study of the impact of radiation quality on cell fate. The observed changes in our tissue-like model after ionizing radiation exposure may explain some discrepancies between radiation-biology studies and clinical data. | | | 26253487
 |
The SMAC mimetic BV6 sensitizes colorectal cancer cells to ionizing radiation by interfering with DNA repair processes and enhancing apoptosis.
Hehlgans, S; Oppermann, J; Reichert, S; Fulda, S; Rödel, C; Rödel, F
Radiation oncology (London, England)
10
198
2015
Abstract anzeigen
In the present study, we aimed to investigate the effect of counteracting inhibitor of apoptosis (IAP) proteins using the small molecule Second Mitochondria-derived Activator of Caspase (SMAC) mimetic BV6 in combination with ionizing radiation on apoptosis, cell cycle regulation, DNA double-strand break (DSB) repair, three-dimensional (3D) clonogenic survival and expression of IAPs in colorectal carcinoma cells.Colorectal cancer cell lines (HCT-15, HT-29, SW480) were subjected to BV6 treatment (0-4 μM) with or without irradiation (2-8 Gy, single dose) followed by MTT, Caspase 3/7 activity, γH2AX/53BP1 foci assays, AnnexinV staining, cell cycle analysis, 3D colony forming assays and Western blotting (cellular IAP1 (cIAP1) and cIAP2, Survivin, X-linked IAP (XIAP)).BV6 treatment decreased cell viability and significantly increased irradiation-induced apoptosis as analyzed by Caspase 3/7 activity, AnnexinV-positive and subG1 phase cells. While basal 3D clonogenic survival was decreased in a cell line-dependent manner, BV6 significantly enhanced cellular radiosensitivity of all cell lines in a concentration-dependent manner and increased the number of radiation-induced γH2AX/53BP1-positive foci. Western blot analysis revealed a markedly reduced cIAP1 expression at 4 h after BV6 treatment in all cell lines, a substantial reduction of XIAP expression in SW480 and HT-29 cells at 24 h and a slightly decreased cIAP2 expression in HCT-15 cells at 48 h after treatment. Moreover, single or double knockdown of cIAP1 and XIAP resulted in significantly increased residual γH2AX/53BP1-positive foci 24 h after 2 Gy and radiosensitization relative to control small interfering RNA (siRNA)-treated cells.The SMAC mimetic BV6 induced apoptosis and hampered DNA damage repair to radiosensitize 3D grown colorectal cancer cells. Our results demonstrate IAP targeting as a promising strategy to counteract radiation resistance of colorectal cancer cells. | | | 26383618
 |
Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method.
Lin, JR; Fallahi-Sichani, M; Sorger, PK
Nature communications
6
8390
2015
Abstract anzeigen
Single-cell analysis reveals aspects of cellular physiology not evident from population-based studies, particularly in the case of highly multiplexed methods such as mass cytometry (CyTOF) able to correlate the levels of multiple signalling, differentiation and cell fate markers. Immunofluorescence (IF) microscopy adds information on cell morphology and the microenvironment that are not obtained using flow-based techniques, but the multiplicity of conventional IF is limited. This has motivated development of imaging methods that require specialized instrumentation, exotic reagents or proprietary protocols that are difficult to reproduce in most laboratories. Here we report a public-domain method for achieving high multiplicity single-cell IF using cyclic immunofluorescence (CycIF), a simple and versatile procedure in which four-colour staining alternates with chemical inactivation of fluorophores to progressively build a multichannel image. Because CycIF uses standard reagents and instrumentation and is no more expensive than conventional IF, it is suitable for high-throughput assays and screening applications. | | | 26399630
 |
Syringaresinol protects against hypoxia/reoxygenation-induced cardiomyocytes injury and death by destabilization of HIF-1α in a FOXO3-dependent mechanism.
Cho, S; Cho, M; Kim, J; Kaeberlein, M; Lee, SJ; Suh, Y
Oncotarget
6
43-55
2015
Abstract anzeigen
Hypoxia-inducible factor 1 (HIF-1) is a master regulator of hypoxic response and has been a prime therapeutic target for ischemia/reperfusion (I/R)-derived myocardial dysfunction and tissue damage. There is also increasing evidence that HIF-1 plays a central role in regulating aging, both through interactions with key longevity factors including Sirtuins and mTOR, as well as by directly promoting longevity in Caenorhabditis elegans.We investigated a novel function and the underlying mechanism of syringaresinol, a lignan compound, in modulation of HIF-1 and protection against cellular damage and death in a cardiomyocyte model of I/R injury. Syringaresinol caused destabilization of HIF-1α following H/R and then protected against hypoxia/reoxygenation (H/R)-induced cellular damage, apoptosis, and mitochondrial dysfunction in a dose-dependent manner. Knock-down of FOXO3 by specific siRNAs completely abolished the ability of syringaresinol to inhibit HIF-1 stabilization and apoptosis caused by H/R. Syringaresinol stimulated the nuclear localization and activity of FOXO3 leading to increased expression of antioxidant genes and decreased levels of reactive oxygen species (ROS) following H/R. Our results provide a new mechanistic insight into a functional role of syringaresinol against H/R-induced cardiomyocyte injury and death. The degradation of HIF-1α through activation of FOXO3 is a potential therapeutic strategy for ischemia-related diseases. | Fluorescence Activated Cell Sorting (FACS) | | 25415049
 |
DNA ligase III acts as a DNA strand break sensor in the cellular orchestration of DNA strand break repair.
Abdou, I; Poirier, GG; Hendzel, MJ; Weinfeld, M
Nucleic acids research
43
875-92
2015
Abstract anzeigen
In the current model of DNA SSBR, PARP1 is regarded as the sensor of single-strand breaks (SSBs). However, biochemical studies have implicated LIG3 as another possible SSB sensor. Using a laser micro-irradiation protocol that predominantly generates SSBs, we were able to demonstrate that PARP1 is dispensable for the accumulation of different single-strand break repair (SSBR) proteins at sites of DNA damage in live cells. Furthermore, we show in live cells for the first time that LIG3 plays a role in mediating the accumulation of the SSBR proteins XRCC1 and PNKP at sites of DNA damage. Importantly, the accumulation of LIG3 at sites of DNA damage did not require the BRCT domain-mediated interaction with XRCC1. We were able to show that the N-terminal ZnF domain of LIG3 plays a key role in the enzyme's SSB sensing function. Finally, we provide cellular evidence that LIG3 and not PARP1 acts as the sensor for DNA damage caused by the topoisomerase I inhibitor, irinotecan. Our results support the existence of a second damage-sensing mechanism in SSBR involving the detection of nicks in the genome by LIG3. | | | 25539916
 |
The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA.
Diner, BA; Li, T; Greco, TM; Crow, MS; Fuesler, JA; Wang, J; Cristea, IM
Molecular systems biology
11
787
2015
Abstract anzeigen
The human PYHIN proteins, AIM2, IFI16, IFIX, and MNDA, are critical regulators of immune response, transcription, apoptosis, and cell cycle. However, their protein interactions and underlying mechanisms remain largely uncharacterized. Here, we provide the interaction network for all PYHIN proteins and define a function in sensing of viral DNA for the previously uncharacterized IFIX protein. By designing a cell-based inducible system and integrating microscopy, immunoaffinity capture, quantitative mass spectrometry, and bioinformatics, we identify over 300 PYHIN interactions reflective of diverse functions, including DNA damage response, transcription regulation, intracellular signaling, and antiviral response. In view of the IFIX interaction with antiviral factors, including nuclear PML bodies, we further characterize IFIX and demonstrate its function in restricting herpesvirus replication. We discover that IFIX detects viral DNA in both the nucleus and cytoplasm, binding foreign DNA via its HIN domain in a sequence-non-specific manner. Furthermore, IFIX contributes to the induction of interferon response. Our results highlight the value of integrative proteomics in deducing protein function and establish IFIX as an antiviral DNA sensor important for mounting immune responses. | | | 25665578
 |
NF-κB signaling mediates acquired resistance after PARP inhibition.
Nakagawa, Y; Sedukhina, AS; Okamoto, N; Nagasawa, S; Suzuki, N; Ohta, T; Hattori, H; Roche-Molina, M; Narváez, AJ; Jeyasekharan, AD; Bernal, JA; Sato, K
Oncotarget
6
3825-39
2015
Abstract anzeigen
PARP inhibitors are a class of promising anti-cancer drugs, with proven activity in BRCA mutant cancers. However, as with other targeted agents, treatment with PARP inhibitors generates acquired resistance within these tumors. The mechanism of this acquired resistance is poorly understood. We established cell lines that are resistant to PARP inhibitor by continuous treatment with the drug, and then used RNA sequencing to compare gene expression. Pathway analysis on the RNA sequencing data indicates that NF-κB signaling is preferentially up-regulated in PARP inhibitor-resistant cells, and that knockdown of core components in NF-κB signaling reverses the sensitivity to PARP inhibitor in resistant cells. Of therapeutic relevance, we show that PARP inhibitor-resistant cells are sensitive to an NF-κB inhibitor in comparison to their parental controls. Malignancies with up-regulation of NF-κB are sensitive to bortezomib, a proteasome inhibitor that is currently used in the clinic. We also show that treatment with bortezomib results in cell death in the PARP inhibitor-resistant cells, but not in parental cells. Therefore we propose that up-regulation of NF-κB signaling is a key mechanism underlying acquired resistance to PARP inhibition, and that NF-κB inhibition, or bortezomib are potentially effective anti-cancer agents after the acquisition of resistance to PARP inhibitors. | | | 25686825
 |
CHK1-driven histone H3.3 serine 31 phosphorylation is important for chromatin maintenance and cell survival in human ALT cancer cells.
Chang, FT; Chan, FL; R McGhie, JD; Udugama, M; Mayne, L; Collas, P; Mann, JR; Wong, LH
Nucleic acids research
43
2603-14
2015
Abstract anzeigen
Human ALT cancers show high mutation rates in ATRX and DAXX. Although it is well known that the absence of ATRX/DAXX disrupts H3.3 deposition at heterochromatin, its impact on H3.3 deposition and post-translational modification in the global genome remains unclear. Here, we explore the dynamics of phosphorylated H3.3 serine 31 (H3.3S31ph) in human ALT cancer cells. While H3.3S31ph is found only at pericentric satellite DNA repeats during mitosis in most somatic human cells, a high level of H3.3S31ph is detected on the entire chromosome in ALT cells, attributable to an elevated CHK1 activity in these cells. Drug inhibition of CHK1 activity during mitosis and expression of mutant H3.3S31A in these ALT cells result in a decrease in H3.3S31ph levels accompanied with increased levels of phosphorylated H2AX serine 139 on chromosome arms and at the telomeres. Furthermore, the inhibition of CHK1 activity in these cells also reduces cell viability. Our findings suggest a novel role of CHK1 as an H3.3S31 kinase, and that CHK1-mediated H3.3S31ph plays an important role in the maintenance of chromatin integrity and cell survival in ALT cancer cells. | | | 25690891
 |